Frequently Asked Questions
NIAGADS and Partners
The National Institute for Aging Genetics of Alzheimer's Disease Data Storage Site (NIAGADS) is the designated data repository for genetic and genomic data generated from NIA grants. NIAGADS also accepts AD genetics and genomics data from studies not funded by the NIA.
NIAGADS is a U24 cooperative agreement between the NIA and the University of Pennsylvania and has been in existence since 2013. In 2017, NIAGADS developed the Data Sharing Service, a FISMA compliant framework that makes it easier for users and administrators to apply for, access, and store AD genetics and genomics data.
Currently, the NIAGADS team is working on harmonizing, standardizing, and moving over legacy datasets from the original NIAGADS database to the DSS service in addition to processing and depositing new datasets for the research community. While the move is taking place, legacy datasets will be available to research on the NIAGADS site. All new datasets are available on the DSS site.
NIAGADS stores genetics and genomics data.
NCRAD stores and distributes biospecimens.
NACC stores and distributes standardized clinical and neuropathological research data for the ADRCs.
AD Knowledge Portal stores the multi-omic and drug development data.
The NIAGADS team is happy to support ID mapping to NACC, NCRAD, and AD Knowledge Portal datasets. If you need help mapping subject IDs, please reach out to help@niagads.org.
NOT-OD-24-157 - Updates to the NIH Genomic Data Sharing Policy
Yes, NIAGADS DSS has obtained the Federal Information Security Management Act (FISMA) Moderate Authority to Operate (ATO) from the University of Pennsylvania School of Medicine. To ensure compliance with federal security standards, the DSS follows the NIST Risk Management Framework (RMF) and adheres to NIST Special Publication (SP) 800-53 Rev4 Moderate Baseline. Transition to the NIST 800-53 Rev5 Moderate Baseline is currently underway for completion in Q2 2025.
All users, US-based and non-US-based, will be required to attest that their institution and any third-party system or Cloud Service Provider involved in data analysis or storage comply with NIST SP 800-171 or an equivalent ISO/IEC 27001/27002 standards as stipulated by the NIH Security Best Practices for Users of Controlled-Access Data. More information about the standard can be found on the NIH website here: NIH-Security-BPs-for-Users-of-Controlled-Access-Data.pdf
As of April 2, 2025, all new Data Access Requests or renewals must attest to the adherence of the NIH Security Best Practices for Users of Controlled-Access Data described in NOT-AD-24-157.
All controlled-access data in NIAGADS falls under the policies set forth in NOT-AD-24-157. This includes any genetic/genomic and non-genetic/genomic data accessed through the qualified access process with DAR approval.
On the Data Access Request (DAR) form, there is a checkbox under the “NIH Genomic Data Sharing Security Attestation” section. Both the investigator requesting data and the Signing Official must attest to the implementation of the policy by checking the “I agree” box on the DAR.
Once your DAR has expired you are expected to close out your project and certify that the dataset(s) available have been destroyed in accordance with the NIH Security Best Practices for Users of Controlled-Access Data unless required to be retained as described in the Data Use Certification Agreement.
Accessing Data in NIAGADS and DSS
To be eligible to access the data investigators must meet the following criteria:
Be permanent employees of their institution at a level equivalent to a full-time assistant, associate, or full professor. Lab staff, trainees (e.g., graduate students) and postdocs are not permitted to submit (NIAGADS and DSS)
Submitting investigators and their signing official must have an eRA Commons ID (DSS only)
This is done through the NIH. Go to http://www.era.nih.gov and click on the accounts tab to learn more.
Only individuals with legal signing authority, can complete this process, see, Registering Institutions (nih.gov).
Yes
Yes
To access data in the DSS you need to complete a Data Access Request (DAR). Information required for a DAR is listed below:
Research Use Statement
Other Project Information
(PI, Institution, IT Director, Signing Officer, Staff and Collaborators, etc)
IRB Approval
IRB Protocol
Data Use Certification
NIAGADS Data Distribution Agreement
NIA Genomic Data Sharing Policy
Derived/Secondary Data Return Plan
Data Transfer Agreement
Cloud Use Statement and Cloud Server Provider Information (if applicable)
Non-Technical Summary
Templates and instructions for filling out each form can be found on here: Application Instructions - DSS NIAGADS
Requests are reviewed by multiple administrators as well as two review committees. Unfortunately, this can make it difficult to estimate total review times. To minimize review times, it is important to submit documents as described in the application instructions on the NIAGADS and DSS Sites.
If requesting data from DSS, be sure to send your signed Data Transfer Agreement (DTA) to niagads@pennmedicine.upenn.edu in a timely manner. Data Access Requests cannot be approved until a fully executed DTA is received from Penn’s contracts department.
If the submission instructions are followed carefully, most applications are approved. Please reach out to the NIAGADS team at niagads@pennmedicine.upenn.edu should you have any questions.
Please make sure you are logging into DARM with your eRA commons ID username and not with your Login.gov credentials. Also be sure you are logged out of login.gov before using your eRA commons ID to log into DARM. If you are still having trouble, you may need to change your eRA commons password. More information on that can be found here: https://www.era.nih.gov/erahelp/Commons/Commons/access/reset_pswd.htm.
No data made available through NIAGADS DSS has an embargo.
Data Types
Click on the dataset tab at the top of the home page. Here you can either use the search bar at the top to enter your query or you can use the filter menu on the side to narrow down results. The lock next to the dataset name tells you if the dataset is publicly available (no application) or restricted access (requires application). Some datasets have publicly available files and restricted files. Within each dataset page you can find details about the study it came from, a breakdown of the participant population, and any related publications.
The Alzheimer’s Genomics Database is also another great resource for this. It has the publicly available GWAS summary statistics available to browse with an easy-to-use interface.
If you are still unsure, you can always email the NIAGADS team at help@niagads.org for assistance.
Yes, you need to specify the dataset in your application.
Downloading Data
NIAGADS incurs the cost for investigators to download most of the available data, including joint genotype-called project level VCFs, phenotypes, and associated meta-data. All files <5Gb can be downloaded directly through the portal, while files >5Gb will need to be downloaded directly from Amazon. In order to download data >5Gb in size, please ensure that you have an Amazon Web Services account. Additional information about using Amazon can be found on our Amazon Instructions page.
CRAMs, gVCFs, and SV VCFs can be downloaded using the Amazon Requester Pays option, which means that the requesting institution will incur the cost of downloading the data.
Options for AWS download using Requestor Pays option:
AWS resource - You would not be charged if you download within the same region as our S3 bucket, US-East (N. Virginia), to another US-East (N. Virginia)
Local download - an affordable transfer option is an Amazon Snowball. DSS would send the data to your S3 bucket, then you can create an AWS Snowball export. The device costs $250 to transfer 80TB of data (plus additional fees.
CRAMs, gVCFs, and SV VCFs can be downloaded using the Amazon Requester Pays option, which means that the requesting institution will incur the cost of downloading the data.
Options for AWS download using Requestor Pays option:
AWS resource - You would not be charged if you download within the same region as our S3 bucket, US-East (N. Virginia), to another US-East (N. Virginia)
Local download - an affordable transfer option is an Amazon Snowball. DSS would send the data to your S3 bucket, then you can create an AWS Snowball export. The device costs $250 to transfer 80TB of data (plus additional fees.
Submitting Data
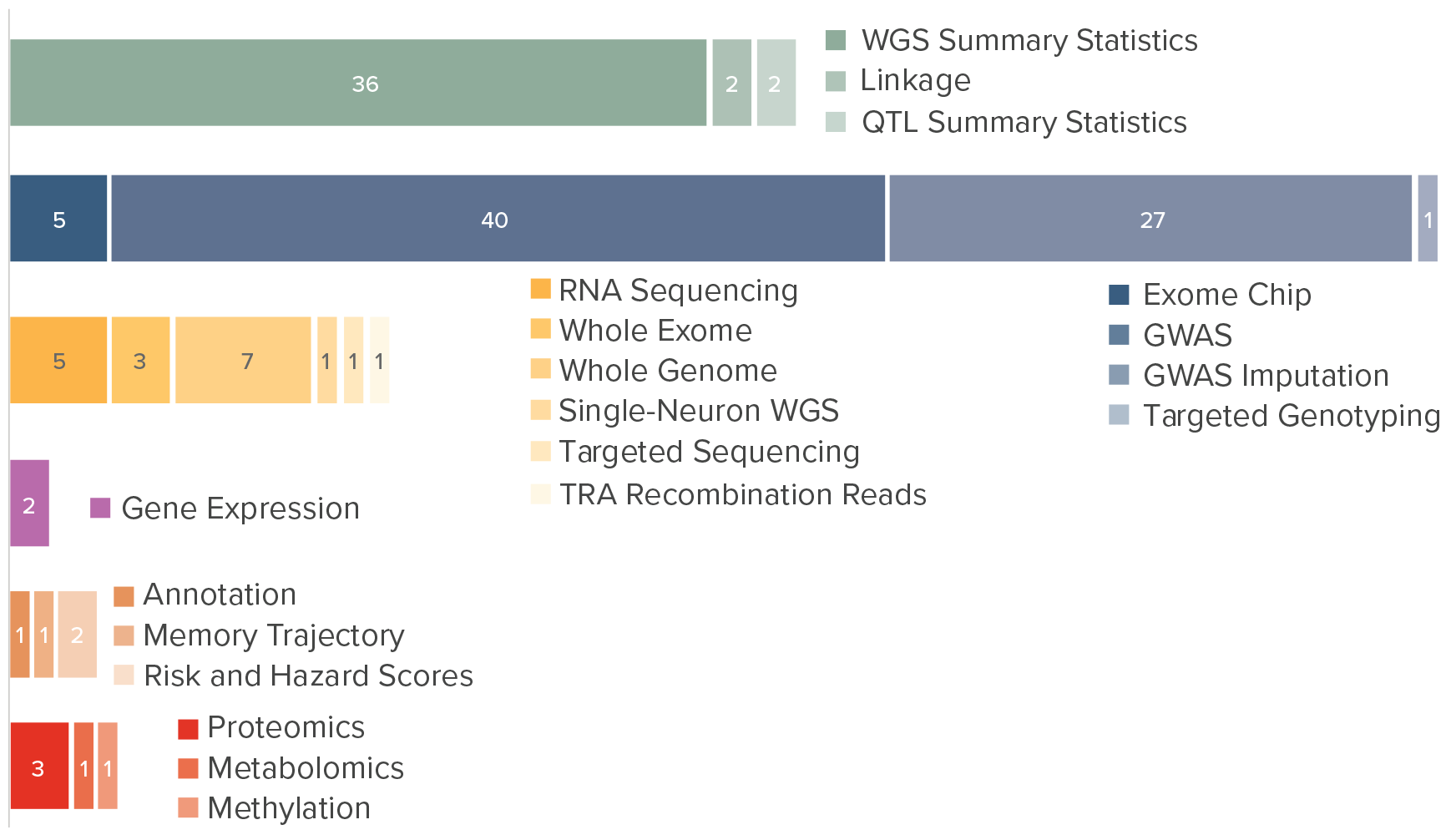
NIAGADS accepts most types of genetics and genomics data. We currently house genotype, GWAS, WGS, WES, RNA-seq, microarray, epigenetic studies (ChIP-seq, ect), WTL, methylation, proteomic, as well as single cell data for RNA-seq, microarray, and epigenetic studies. Please see the data submission instructions for a list of datatypes and documentation requirements here: Data Submission Instructions - DSS NIAGADS
We are always open to taking in new types of genetics and genomics data. Contact help@niagads.org if you have questions about data type submission for existing or new data types.
To deposit data, please email help@niagads.org and review the data submission instructions page for the additional documentation needed to support deposition of your dataset.
Please see the data submission instructions page for the additional documentation needed to support deposition of your dataset.
The NIAGADS team will work with you to transfer your data over an FTP link or other cloud service. Please contact help@niagads.org to coordinate.
Please see NIAGADS Data Storage Cost Estimates for appliable fees for storage at NIAGADS. If you plan to submit a dataset over 50TB or >1K short-read raw WGS samples, please reach out to help@niagads.org to discuss storage and transfer options.
This depends on the receipt of all the documentation needed for submission in a timely manner. Once we have all documentation, and it is complete, it takes about 2 months to deposit the dataset and make it available.
Yes, you can always update your data set. To update your data set, please contact help@niagads.org with the existing accession number and a mapping of which files should replace existing files, are new, or should no longer be shared.
Datasets made available via controlled-access are available for request by qualified investigators with an eRA Commons account. They must have IRB approval and their research use is reviewed and approved by the NIAGADS ADRD Data Access Committee (NADAC), made up of program officials from the NIH. Yearly renewal of their approved data access request is required in order to maintain access to the data.
Datasets that do not required controlled access and are available for public access (e.g. p-values) are made freely and openly available through our Open Access Data Portal.
Alzheimer’s Disease Sequencing Project (ADSP)
For the latest information about the data available from the ADSP visit https://adsp.niagads.org/data/data-summary/
Support and Resources
NIAGADS currently offers extensive documentation on applying, accessing and submitting data here: Data Application and Submission - DSS NIAGADS
Video tutorials can be found on our YouTube site here: NIAGADS - YouTube
Lastly, we hold office hours throughout the year, anyone can join. Subscribe to our mailing list to be notified when the next office hour will occur
Yes, they can be found here: NIAGADS - YouTube
You can always reach out to help@niagads.org if you have any questions and a member of our team will assist you.
We also hold office hours throughout the year with different subject matter experts from our team to answer questions live. If you would like to be kept informed of future office hours, please opt into our mailing list here
NIAGADS currently offer four tools for users.
| ||||
Description | Contains curated ADRD variants from the literature | Contains functional genomics and annotation data for GRCh37/g19 and GRch38/hg38 | Browse NIAGADS GWAS summary statistics and ADSP variant annotations or view summaries in the context of genes and variants | Variant browser
Browse for ADSP variants instead of downloading the VCFs |
What does it cover? | Variants specific to AD | Whole genome | Whole genome with emphasis on contextualizing AD | Specific to ADSP dataset (R1-R4) |
Data Types | Alzheimer’s disease variants and supporting literature | Functional genomic data such as tissue/cell-type specific epigenetic marks, QTL, chromatin interaction, TF binding. | Genes and variants across the whole genome
NIAGADS publicly available GWAS summary statistics | ADSP variants of significance (R1-R4) |
Use it for | Look up and review supporting information for high confidence AD variants | Use the harmonized genomic and annotation data collections for downstream genetic and genomic analyses such as GWAS variant analysis, genomic regions and other analyses. | View gene and variant reports to quickly discover existing evidence for AD-association or use the NIAGADS genome browser to explore AD GWAS summary statistics in a broader genomic context. | Open access, querying variants in the ADSP data without downloading VCFs |
See the tools section below for additional frequently asked questions.
Tools
Alzheimer’s Disease Variant Portal (ADVP)
The Alzheimer’s Disease Variant Portal is a database of literature derived populations specific variants associated with AD collated form published GWAS papers from the Alzheimer’s Disease Genetics Consortium (ADGC) and other consortiums.
Publication where genome-wide significant and suggestive loci from AD genetic studies (ADGC and other AD GWAS studies in the NHGRI/EBI GWAS catalog) were selected if they reported GWAS finding. From these selected publications, only main/major findings were extracted, and then a systemic data extraction and curation procedure for each publication was applied. Data was then harmonized to ensure consistency and reduce variability when reporting information (e.g., CSF AB1-42 or CSF Abeta would be the same and reported as CSF ab1-42). A second curator validated the work of the first curator, and lastly extracted data was parsed customized scripts to validate, annotate, and store the publication, variant, and association data in the relational database. For detail information on the curation process, see the associated publication here.
V1.0 (2/21) contains >900 loci, >1,800 variants, >80 cohorts, and 8 populations cataloging 6,990 associations related to disease risk, expression quantitative trails, endophenotypes, and neuropathy. Data primarily comes from studies conducted by the Alzheimer’s Disease Genetics Consortium (ADGC).
Investigator can use the ADVP to quickly and systematically explore high-confidence AD genetics findings and review insights into population specific AD genetic architecture.
Alzheimer’s Genomics Database
The Alzheimer’s Genomics Database is a user-friendly web-knowledgebase for searching annotated AD-relevant GWAS summary statistics datasets. It also facilitates real-time data mining and provides summaries of association records in the context of functionally annotated genes and variants. Interactive visualizations, such as the NIAGADS Genome Browser allow users to explore these data tracks and annotations in the broader genomic context and compare against functional genomics tracks from FILER.
56 publicly summary statistics deposited in NIAGADS are available, as well as ~263 million annotated variants (including 232M from the ADSP, ~296K with significant AD/ADRD-risk association, and ~20K annotated genes. For more details about the data available visit the documentation page: GDB |About (niagads.org).
Quickly search a gene or variant to see supporting evidence from the publicly available GWAS summary statistics deposited in NIAGADS and link to other resources, like AD Knowledge Portal and UniProtKB to explore additional evidence. You can also examine the datasets with publicly available summary statistics to help determine if you would like to submit a Data Access Request (DAR) for a particular dataset.
See our YouTube tutorials for navigating the site here: Navigating the NIAGADS Alzheimer's Genomics Database - YouTube
FILER
FILER is a functional genomics database with a large curated and integrated collection of harmonized, extensible, indexed, and searchable human functional genomics data from >20 data sources.
>50,000 harmonized, annotated genomic datasets across >20 integrate data sources, >1,100 tissues/cell types and >20 experimental assays spanning 17 billion genomic records on GRch37/hg19 and GRCh38/hg38. To learn more about the datatypes available, click here.
Primary annotated and functional genomics datasets from existing data sources are collected and complied into a unified catalogue. Then individual genomic datasets are curated, processed, and imported into FILER using the FILER data harmonization and annotation pipeline. Data type specific metadata was then harmonized to generate standardized consistent metadata descriptions of each of the filer data tracks (sets of genomic/annotation records). To learn more about the curation process, please check out the information here.
Use the harmonized genomic and annotation data collect for downstream genetic and genomic analysis, further interpretation, characterization, and discovery for GWAS results and biological experiments.
Yes. You can use FILER to find overlaps with your data (in the form of genomic intervals) from other types of user experiments like ChIP-seq peaks, small RNA peaks, or ATAC-seq open chromatin regions.
FILER can be deployed two ways:
Locally on a server or cluster
Through the webserver
To learn more about how to use FILER, including it API feature, visit the about page, where there is a user tutorial as well as other support information.
VariXam
VariXam is an aggregated database of genomic variants detected in data via whole-genome/whole-exome sequencing from the Alzheimer’s Disease Sequencing Project (ADSP). VariXam is also comprised of a variant browser to visualize this data.
The database can also be accessed through command line. Visit the VariXam website to learn more.
Currently the database includes variants from the R1 5K WGS, R2 20K WES, R3 17K WGS, and R4 36K WGS. The human reference genome used is GRCh38.
Variants were processed by the Variant Calling Pipeline and data management tool (VCPA). The VCPA consists of two independent but linkable components.
The pipeline – details about the pipeline can be found in the link above.
The tracking database
Quickly search for variants of interest in the ADSP to see variants of significance for AD and related dementias without the need to apply for the full dataset or download and process VCFs.
Type your gene, variant, position, or refSNP id into the search bar and click enter. Variant records from the ADSP dataset will be returned in a table below the search bar.
There is also an API which can be used to query the variables above. For more details about the API, visit the VariXam website.
NIAGADS (API)
The NIAGADS Application Programming Interface (API) is a public resource allowing programmatic access the publicly available data stored in NIAGADS knowledgebases. An alpha version is now available, allowing basic queries against the ADVP, GenomicsDB, and FILER to retrieve data track metadata, track hits within a genomic span of interest, and genomic evidence for AD-risk association for genes and variants. It is a user-friendly framework for extracting information and integrating AD-relevant gene, variant, and functional genomic annotations into analysis pipelines.
The NIAGADS API is an intuitive, templated RESTful interface for programmatic querying that meets OpenAPI standards. Endpoints are simple URLs that specify the knowledgebase, feature type (gene, variant), feature identifier (s) and filter criteria (experiment or bio-sample properties).
Use of the API requires only a HTTPS library and JSON parser, permitting largescale programmatic access in most programming languages.
Check out the API at http://api.niagads.org/ to learn more.
